Quanjiang Ji Group
| Principal Investigator |
|
| Research |
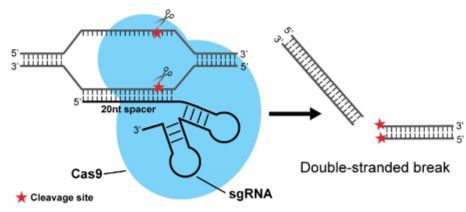
The emergence of drug-resistant human pathogens has posed a severe public crisis worldwide. To counter infections caused by major human pathogens, we aim to create novel genome editing tools, address fundamental infection and drug-resistant mechanisms, and develop new therapeutic means with the utilization of multiple approaches (chemical, biological, and engineering). Genome editing in human pathogens: Genetics is the key means to study bacterial physiology. However, traditional genetic manipulation methods in major human pathogens remain as time-consuming and laborious endeavors. We have created rapid and highly efficient genetic manipulation tools in multiple major human pathogens, including Staphylococcus aureus (JACS, 2017; Chem Sci, 2018), Pseudomonas aeruginosa (iScience, 2018), and Klebsiella pneumoniae (Appl Environ Microbiol, 2018) by engineering the powerful CRISPR/Cas9 genome editing technology and deaminase-mediated base editing systems. These tools have been requested and utilized by numerous research groups worldwide and are/will be available in Addgene (http://www.addgene.org/Quanjiang_Ji/). We utilize protein engineering and synthetic biology approaches to develop genome-wide screening tools and aim to construct label-free gene-knockout libraries in multiple human pathogens. The development of these tools will advance fundamental physiology studies as well as novel drug-target exploration. We study fundamental DNA recognition and cleavage mechanisms of CRISPR systems. We design, engineer, and functionalize CRISPR systems for diverse applications, in particular in the study of infectious diseases.
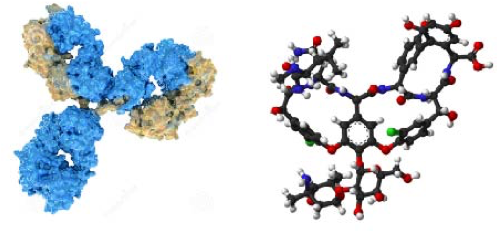
Fundamental infection and drug-resistant mechanisms: We study basic infection and drug-resistant mechanisms in major human pathogens by taking advantages of structural biology and genome editing approaches (PNAS, 2018; Mol Microbiol, 2018). Recently, we focused on elucidating the molecular mechanisms of nicotianamine-like metallophore-mediated transition metal acquisition processes as well as biofilm-formation mechanisms of several cell surface proteins in human pathogens. We are also interested in small-molecule signaling and regulatory pathways that affect bacterial pathogenesis and drug resistance.
Therapeutic means against infections: We develop therapeutic antibodies and small molecules targeting key virulence or drug-resistant proteins, in particular the extracellular proteins that play vital roles in bacterial metal acquisition and biofilm formation. We aim to identify new drug targets using the mutant libraries we are currently constructing and screen effective antibodies or small molecules against them.
|
| Publications |
2018 6. Wang, Y., Wang, S., Chen, W., Song, L., Shen, Z., Yu, F., Li, M., Ji, Q.*(2018) Precise and efficient genome editing in Klebsiella pneumoniaeusing CRISPR-Cas9 and CRISPR-assisted cytidine deaminase. Appl Environ Microbiol DOI: 10.1128/AEM.01834-18 5. Chen, W., Zhang, Y., Zhang, Y., Pi, Y., Gu, T., Song, L., Wang, Y., Ji, Q.* (2018) CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. iScience 6: 222-31. 4. Wei, W.#, Zhang, Y.#, Gao, R., Li, J., Xu, Y., Wang, S., Ji, Q.*, Feng, Y.* (2018) Crystal structure and acetylation of BioQ suggests a novel regulatory switch for biotin biosynthesis in Mycobacterium smegmatis. Mol Microbiol DOI: 10.1111/mmi.14066. 3. Song, L., Zhang, Y., Chen, W., Gu, T., Zhang, S.Y., Ji, Q.* (2018) Mechanistic insights into staphylopine-mediated metal acquisition. Proc Natl Acad Sci U S A 115: 3942-7. 2. Gu, T.#, Zhao, S.#, Pi, Y., Chen, W., Chen, C., Liu, Q., Li, M., Han, D.*, Ji, Q.* (2018) Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci9: 3248-53. 2017 1. Chen, W., Zhang, Y., Yeo, W.S., Bae, T., Ji, Q.* (2017) Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J Am Chem Soc 139: 3790-5. PRIOR TO SHANGHAITECH
|
| Patents |
6. 季泉江、王宇。一种用于肺炎克雷伯菌基因编辑的双质粒系统。申请号:201811039504.9 5. 季泉江、王宇。一种肺炎克雷伯菌基因编辑的表达载体。申请号:201811039489.8 4. 季泉江、陈未中。一种pnCasPA-BEC质粒及其应用。申请号:201810767194.6 3. 季泉江、陈未中。一种pCasPA/pACRISPR双质粒系统及其应用。申请号:201810766759.9 2. 季泉江、顾桐年。一种pnCasSA-BEC质粒及其应用。申请号:201810169946.9 1. 季泉江、陈未中。一种pCasSA质粒及其应用。申请号:201611255504.3 |
| Group Activities |
|
| Current Group Members |












| Former Group Members |